Download Manager
Overview
Download Manager is a utility software from Illumina Connected Annotation software suite for user to easily pick and choose prebuilt annotation databases compatible with Illumina Connected Annotation.
Previously, we provided the Downloader software that the user can run to download the latest annotation database automatically. With the new Download Manager, users can choose the exact version of an annotation database they want.
Prerequisite
Creating Illumina API key
To use Download Manager, users have to create an Illumina API key. To generate the API key, users are required to create Illumina public account which can be created freely from this page.
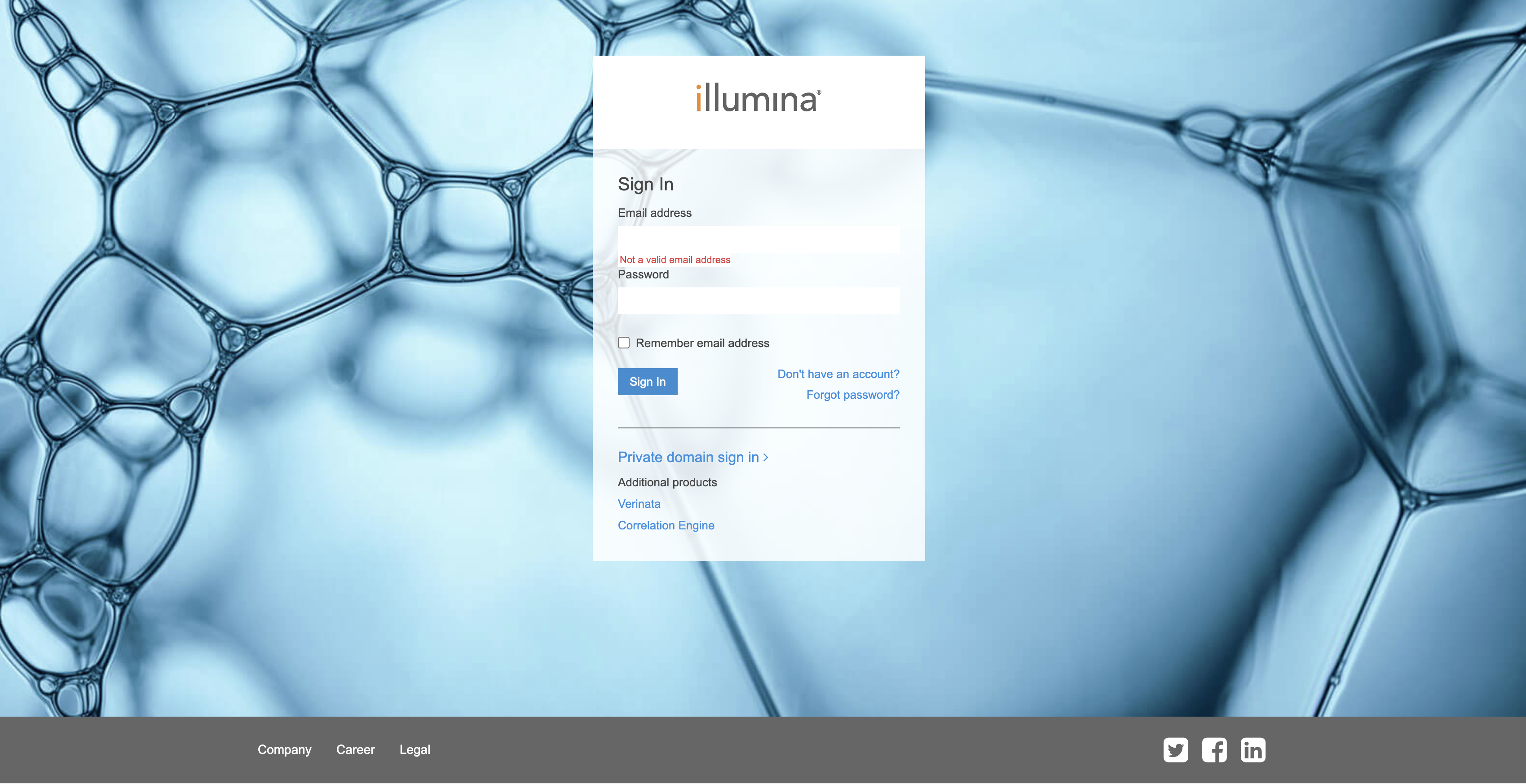
Click "Don't have an account" link to go to the user creation form. If you already have an account, enter your credentials. Once you login, you will go to Product Dashboard page. Click on the user icon on the top right to open menu and choose Manage API Keys.
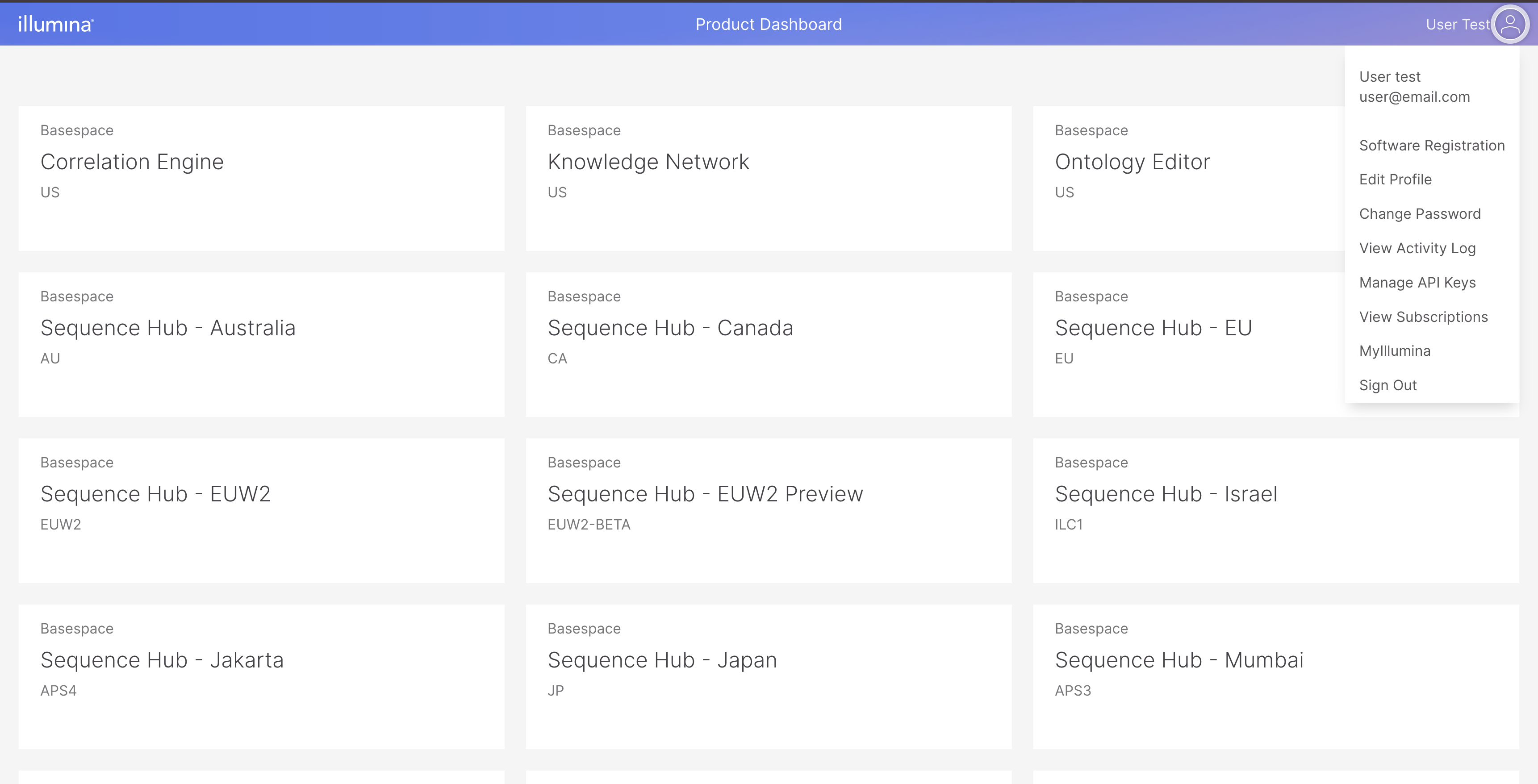
In the Manage API Keys, click Generate Button and enter the API key name. Once you have done that, the generated API key will be displayed.
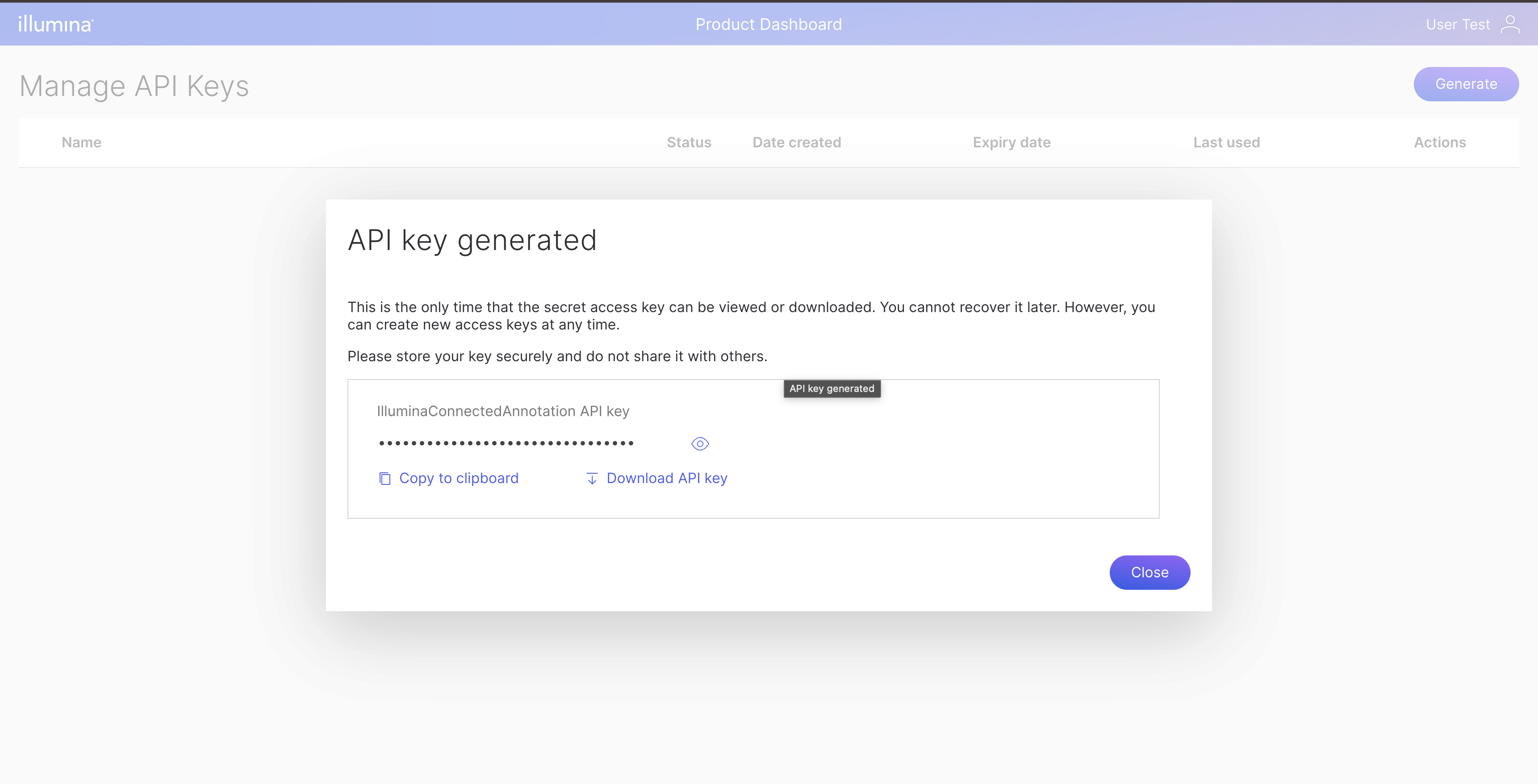
Storing Illumina API key in your system
Copy the API key and store it into a file with json format like below:
{
"IlmnApiKey": "<your api key>"
}
By default, Download Manager utility will look for file ~/.ilmnAnnotations/credentials.json but you can also save the API key in different path and provide it as command line parameter later.
If you have API key and API secret for Illumina Connected Annotation professional tier data source, you should put those credentials in this file also. Below is the format if you have professional data source API key and API secret:
{
"IlmnApiKey": "<your Illumina account api key>",
"ApiKey": "<your professional data source API key>",
"ApiSecret": "<your professional data source API secret>"
}
Commands
There are several commands in Download Manager utility.
List
The first command is list. This command is to list all available data annotation and version.
dotnet DownloadManager.dll list
---------------------------------------------------------------------------
DownloadManager (c) 2024 Illumina, Inc.
3.24.0
---------------------------------------------------------------------------
USAGE: dotnet DownloadManager.dll list [options]
List available data
OPTIONS:
--credential, -l <VALUE>
Path to credential file. The default file
location is ~/.ilmnAnnotations/credentials.json
--ref, -r <VALUE> Assembly reference (GRCh38/GRCh37)
--dir, -d <VALUE> Local directory
--sources, -s <VALUE> data source to show
--help, -h displays the help menu
--version, -v displays the version
##### Supported Annotation Sources #####
Basic Tier: DECIPHER, GME, GERP, DANN, REVEL, ClinGen, gnomAD, phyloP, TOPMed, DGV, 1000 Genomes, CliinVar, dbSNP, FusionCatcher, MITOMAP, MultiZ100Way
Professional Tier: PrimateAI(GRCh37), PrimateAI-3D(GRCh38), SpliceAI, COSMIC, OMIM.
##### Contact #####
Professional content licensing, feedback and technical support: annotation_support@illumina.com.
There are several parameters that you can pass.
- Param
--refor-ris a required parameter. This is to filter data based on genome assembly reference. The value is either GRCh37 or GRCh38 - Param
--diror-dis an optional parameter. This param is the path to your local data directory. If you provide this param, the list will mark file that you have already downloaded. - Param
--credentialor-lis an optional parameter. This param is the path to your credential file. - Param
--sourcesor-sis a filter based on data source. Without this param, the list will display all data source but only display the latest version available. If you filter using this param, you will see all available version.
Below are some example of the output:
dotnet DownloadManager.dll list -r GRCh38 -d /path/to/folder
DownloadManager (c) 2024 Illumina, Inc.
3.24.0
---------------------------------------------------------------------------
======================================================================================================================================================
Data Source | Annotation Type | Description | Version
======================================================================================================================================================
DANN | Score | DANN | 20200205 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
DECIPHER | StructuralVariant | DECIPHER | 201509 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
Ensembl | GeneModels | Ensembl | 110 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
FusionCatcher | GeneFusion | FusionCatcher | 1.33 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
GME | SmallVariant | GME | 20160618 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
GenomeAssembly | GenomeAssembly | GenomeAssembly | GRCh38.p12 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
Gerp | Score | Gerp | 20110522 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
HGNC | GeneModels | HGNC | 20230824 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
MultiZ100Way | Protein | MultiZ100Way | 20171006 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
PrimateAI | SmallVariant | PrimateAI | 0.2 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
REVEL | SmallVariant | REVEL | 20200205 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
RefSeq | GeneModels | RefSeq | GCF_000001405.40-RS_2023_03 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
TOPMed | SmallVariant | TOPMed | freeze_5 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
clingen | Gene | ClinGen disease validity curations | 20240508 (*)
| | ClinGen Dosage Sensitivity Map | 20240508 (*)
| StructuralVariant | ClinGen Dosage Sensitivity Map | 20240508 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
clingen (legacy) | StructuralVariant | ClinGen | 20160414 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
clinvar | SmallVariant | ClinVar | 20240502 (*)
| StructuralVariant | ClinVar | 20240502 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
clinvar-preview | SmallVariant | ClinVarPreview | 20240502 (*)
| StructuralVariant | ClinVarPreview | 20240502 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
cosmic | Gene | Cosmic Cancer Gene Census | 99 (*)
| GeneFusion | COSMIC gene fusions | 99 (*)
| SmallVariant | COSMIC | 99 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
dbSNP | SmallVariant | dbSNP | 156 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
globalAllele | SmallVariant | dbSNP | 151 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
gnomad | Gene | gnomAD_gene_scores | 4.0 (*)
| LowComplexityRegions | gnomAD_LCR | 2.1 (*)
| SmallVariant | gnomAD | 4.1 (*)
| StructuralVariant | gnomAD_SV | 4.0 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
gnomad-exome | SmallVariant | gnomAD_exome | 4.1 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
mitomap | SmallVariant | MITOMAP | 20200819 (*)
| StructuralVariant | MITOMAP_SV | 20200819 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
omim | Gene | OMIM | 20240508 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
oneKg | RefMinor | 1000 Genomes Project | Phase 3 v3plus (*)
| SmallVariant | 1000 Genomes Project | Phase 3 v3plus (*)
| StructuralVariant | 1000 Genomes Project (SV) | Phase 3 v5a (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
phylopScore | ConservationScore | phyloP | hg38 (*)
| Score | PhyloPPrimate | 1.0 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
primateAI-3D | SmallVariant | PrimateAI-3D | 1.0 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
spliceAI | SmallVariant | SpliceAI | 1.3 (*)
------------------------------------------------------------------------------------------------------------------------------------------------------
(*) : available in local directory
dotnet DownloadManager.dll list -r GRCh38 -d /path/to/folder -s clinvar
---------------------------------------------------------------------------
DownloadManager (c) 2024 Illumina, Inc.
3.24.0
---------------------------------------------------------------------------
======================================================================================================================================================
Data Source | Annotation Type | Description | Version
======================================================================================================================================================
clinvar | SmallVariant | ClinVar | 20240502 (*)
| | ClinVar | 20231230
| | ClinVar | 20231203
| | ClinVar | 20231028
| | ClinVar | 20230930
| | ClinVar | 20230903
| | ClinVar | 20230822
| | ClinVar | 20230608
| | ClinVar | 20230509
| | ClinVar | 20230301
| | ClinVar | 20230210
| | ClinVar | 20220901
| | ClinVar | 20220505
| | ClinVar | 20211202
| | ClinVar | 20210805
| | ClinVar | 20210603
| | ClinVar | 20210401
| | ClinVar | 20200903
| | ClinVar | 20200302
| | ClinVar | 20200102
| | ClinVar | 20191001
| | ClinVar | 20190808
| | ClinVar | 20190204
| | ClinVar | 20180129
| StructuralVariant | ClinVar | 20240502 (*)
| | ClinVar | 20231230
| | ClinVar | 20231203
| | ClinVar | 20231028
| | ClinVar | 20230930
| | ClinVar | 20230903
| | ClinVar | 20230822
| | ClinVar | 20230608
| | ClinVar | 20230509
| | ClinVar | 20230301
| | ClinVar | 20230210
| | ClinVar | 20220901
| | ClinVar | 20220505
------------------------------------------------------------------------------------------------------------------------------------------------------
(*) : available in local directory
Create config
The second command is make-config. This command will generate a config file that will be used to define which annotation data and which version to download.
The generated config file is basically a template which you can edit manually. It will contain latest version of all data source. In case you want to exclude some data source or require a different version, you can edit it manually.
dotnet DownloadManager.dll make-config
---------------------------------------------------------------------------
DownloadManager (c) 2024 Illumina, Inc.
3.24.0
---------------------------------------------------------------------------
USAGE: dotnet DownloadManager.dll make-config [options]
Create a downloader config file
OPTIONS:
--credential, -l <VALUE>
Path to credential file. The default file
location is ~/.ilmnAnnotations/credentials.json
--ref, -r <VALUE> Assembly reference (GRCh38/GRCh37)
--out, -o <assembly> Output file name. Default value is ~/.
ilmnAnnotation/assembly_annotation_config.json.
--help, -h displays the help menu
--version, -v displays the version
##### Supported Annotation Sources #####
Basic Tier: DECIPHER, GME, GERP, DANN, REVEL, ClinGen, gnomAD, phyloP, TOPMed, DGV, 1000 Genomes, CliinVar, dbSNP, FusionCatcher, MITOMAP, MultiZ100Way
Professional Tier: PrimateAI(GRCh37), PrimateAI-3D(GRCh38), SpliceAI, COSMIC, OMIM.
##### Contact #####
Professional content licensing, feedback and technical support: annotation_support@illumina.com.
There are several parameters for make-config command.
- Param
--credentialor-lis an optional parameter. This param is the path to your credential file. - Param
--refor-ris a required parameter. This is to filter data based on genome assembly reference. The value is either GRCh37 or GRCh38 - Param
--outor-ois an optional parameter. This param is the path where the config file will be written to. If not specified, it will write to ~/.ilmnAnnotation/{assembly}_annotation_config.json.
Below is example of the generated config file.
{
"Ensembl": {
"GeneModels": "110"
},
"RefSeq": {
"GeneModels": "GCF_000001405.40-RS_2023_03"
},
"GenomeAssembly": {
"GenomeAssembly": "GRCh38.p12"
},
"clinvar": {
"SmallVariant": "20240502",
"StructuralVariant": "20240502"
},
...
}
Download file
The third command available is download. This command will download the database files. Files that already exist in the output folder will be skipped (if the md5checksum match). You may think of this more like a sync command than a download or copy.
It will also auto update if you have older version of the data by deleting the old version and downloading the new version.
dotnet DownloadManager.dll download
---------------------------------------------------------------------------
DownloadManager (c) 2024 Illumina, Inc.
3.24.0
---------------------------------------------------------------------------
USAGE: dotnet DownloadManager.dll download [options]
Download file
OPTIONS:
--credential, -l <VALUE>
Path to credential file. The default file
location is ~/.ilmnAnnotations/credentials.json
--ref, -r <VALUE> Assembly reference (GRCh38/GRCh37)
--config, -c <VALUE> Download file config. By default, it will use
file ~/.ilmnAnnotation/GRChXX_annotation_config.
json
--dir, -d <VALUE> Download file directory
--thread, -t <VALUE> Number of concurrent download
--help, -h displays the help menu
--version, -v displays the version
##### Supported Annotation Sources #####
Basic Tier: DECIPHER, GME, GERP, DANN, REVEL, ClinGen, gnomAD, phyloP, TOPMed, DGV, 1000 Genomes, CliinVar, dbSNP, FusionCatcher, MITOMAP, MultiZ100Way
Professional Tier: PrimateAI(GRCh37), PrimateAI-3D(GRCh38), SpliceAI, COSMIC, OMIM.
##### Contact #####
Professional content licensing, feedback and technical support: annotation_support@illumina.com.
There are several parameters for the download command.
- Param
--credentialor-lis an optional parameter. This param is the path to your credential file. - Param
--refor-ris a required parameter. This is to filter data based on genome assembly reference. The value is either GRCh37 or GRCh38 - Param
--configor-cis an optional parameter. This is the file config that contains database and version that you want to download. If not specified, it will use ~/.ilmnAnnotation/GRChXX_annotation_config. json - Param
--diror-dis a required pqrameter. It specified the directory that will be used to store all downloaded files. - Param
--threador-tis an optional parameter. It is the number of thread that will be used to download file simultaneously. By default, the value is 4.
Below is an example where you download new release of reference file and Ensembl and RefSeq cache. Tge downloader will first detect that you have an existing reference and cache files with different version. The downloader then will delete those files and download the new files.
dotnet DownloadManager.dll download -r GRCh38 -t 8 -d /target/directory
---------------------------------------------------------------------------
DownloadManager (c) 2024 Illumina, Inc.
3.24.0
---------------------------------------------------------------------------
Listing annotation files in local directory...
- File .DS_Store is not supported, skipping file
Requesting remote file information to be downloaded...
Remote file list received!
Syncing local files with requested files
Deleting file /target/directory/Cache/GRCh38.Ensembl.ndb...
Deleting file /target/directory/Cache/GRCh38.Ensembl.ndb.idx...
Deleting file /target/directory/References/Homo_sapiens.GRCh38.Nirvana.dat...
Deleting file /target/directory/Cache/GeneSymbols.ndb...
Deleting file /target/directory/Cache/GeneSymbols.ndb.version.json...
Deleting file /target/directory/Cache/GRCh38.RefSeq.ndb...
Deleting file /target/directory/Cache/GRCh38.RefSeq.ndb.idx...
Requesting remote file information to be downloaded...
Downloading GRCh38.Ensembl.ndb (92.81 MB)
Downloading index file GRCh38.Ensembl.ndb (17.01 KB)
Downloading Homo_sapiens.GRCh38.Nirvana.dat (824.89 MB)
Downloading GeneSymbols.ndb (137.12 KB)
Downloading index file GeneSymbols.ndb (0.15 KB)
Downloading GRCh38.RefSeq.ndb (59.5 MB)
Downloading index file GRCh38.RefSeq.ndb (16.51 KB)
Finish downloading file GeneSymbols.ndb
Finish downloading index file GeneSymbols.ndb
Finish downloading index file GRCh38.Ensembl.ndb
Finish downloading index file GRCh38.RefSeq.ndb
Finish downloading file GRCh38.RefSeq.ndb
Finish downloading file GRCh38.Ensembl.ndb
Finish downloading file Homo_sapiens.GRCh38.Nirvana.dat
Download all files finished!
Below is an example where one of your file is incomplete. In this case, there is no index file. The downloader will detect this and delete the file and redownload both the SA file and its index.
dotnet DownloadManager.dll download -r GRCh38 -t 8 -d /target/directory
---------------------------------------------------------------------------
DownloadManager (c) 2024 Illumina, Inc.
3.24.0
---------------------------------------------------------------------------
Listing annotation files in local directory...
- File .DS_Store is not supported, skipping file
- Error reading local file. File or index file not found for ClinVar_20240502.nsa. Deleting ClinVar_20240502.nsa
Requesting remote file information to be downloaded...
Remote file list received!
Syncing local files with requested files
Requesting remote file information to be downloaded...
Downloading ClinVar_20240502.nsa (95.17 MB)
Downloading index file ClinVar_20240502.nsa (3.73 KB)
Finish downloading index file ClinVar_20240502.nsa
Finish downloading file ClinVar_20240502.nsa
Download all files finished!